Detection of Hantavirus during the COVID-19 Pandemic, Arizona, USA, 2020
Gavriella Hecht
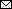
, Ariella P. Dale, Irene Ruberto, Guillermo Adame, Ryan Close, Sarah-Jean Snyder, Kathryn Pink, Nathanael Lemmon, Jessica Rudolfo, Michael Madsen, Andrea L. Wiens, Caitlin Cossaboom, Trevor Shoemaker, Mary J. Choi, Deborah Cannon, Inna Krapiunaya, Shannon Whitmer, Melissa Mobley, Emir Talundzic, John D. Klena, and Heather Venkat
Author affiliations: Arizona Department of Health Services, Phoenix, Arizona, USA (G. Hecht, A.P. Dale, I. Ruberto, G. Adame, H. Venkat); Centers for Disease Control and Prevention, Atlanta, Georgia, USA (A.P. Dale, C. Cossaboom, T. Shoemaker, M.J. Choi, D. Cannon, I. Krapiunaya, S. Whitmer, M. Mobley, E. Talundzic, J.D. Klena, H. Venkat); Indian Health Service, Whiteriver, Arizona, USA (R. Close, K. Pink, N. Lemmon); Indian Health Service, Show Low, Arizona, USA (S.-J. Snyder); White Mountain Apache Tribe, Whiteriver (J. Rudolfo); Coconino County Health and Human Services, Flagstaff, Arizona, USA (M. Madsen); Maricopa County Office of the Medical Examiner, Phoenix (A.L. Wiens)
Main Article
Figure 2

Figure 2. Phylogenetic tree for Orthohantavirus short (S) segment of samples from 2 patients who died of hantavirus infection, Arizona, USA. We inferred the phylogenetic history of full-length Sin Nombre virus S segment using maximum-likelihood estimation. Non–Sin Nombre virus species, Black Creek Canal virus, and Bayou virus are included as outgroups. Bold indicates isolates from this study; squares indicate those from patient 1 and circles those from patient 2. Numbers at nodes indicate bootstrap support >70% after 1,000 iterations. Phylogenetic trees were made using a nucleotide alignment of Orthohantavirus S segments. GenBank accession numbers are provided. Scale bar indicates nucleotide substitutions per site. Additional phylogenetic trees for Orthohantavirus medium and large segments of Sin Nombre virus are in the Appendix.
Main Article
Page created: June 29, 2023
Page updated: July 20, 2023
Page reviewed: July 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
