Volume 29, Number 7—July 2023
Dispatch
Candida vulturna Outbreak Caused by Cluster of Multidrug-Resistant Strains, China
Abstract
Candida vulturna belongs to the Candida haemulonii species complex and is phylogenetically related to C. auris. We report a C. vulturna outbreak among persons in Shanxi Province, China, during 2019–2022. Isolates were resistant to multiple antifungal drugs and exhibited enhanced adhesion and biofilm formation properties.
Candida vulturna, a fungal pathogen that is phylogenetically related to C. haemulonii and C. auris, was isolated from flowers in a taxonomic study of yeasts in 2016 (1,2). Since then, C. vulturna has been sporadically isolated in different countries from clinical specimens such as blood, wounds, and peripherally inserted central catheters (PICCs) (1–4). C. vulturna, C. haemulonii, and C. auris belong to the Metschnikowia/Candida clade (1,5). Antifungal drug resistance, especially to the azoles, is a common feature of species within this clade. During 2009–2022, fungal infections caused by the reportedly rare species C. haemulonii and C. auris have become more prevalent in clinical settings (1,6–10). The increased occurrence of those infections could be the result of the widespread use of antifungal agents in clinical and agricultural settings, as well as the environmental changes caused by human activities (10–12).
In China, reports of infections caused by the superbug fungus C. auris have been relatively infrequent; however, the prevalence of C. haemulonii and associated species in the C. haemulonii complex has been steadily increasing in recent years (8,13). For our study, we analyzed deidentified health records of patients infected with C. vulturna, as approved by the ethics committee of a general hospital in Shanxi Province, China.
We selected a total of 19 patients, 17 male and 2 female, who had been infected with C. vulturna during January 1, 2019–October 26, 2022 (Appendix Figure 1). We isolated 16 C. vulturna strains directly from the blood through venipuncture and 7 strains from a PICC line tip of the 19 patients (Appendix Table). We initially identified the strains as C. haemulonii complex species by growth on CHROMagar Candida medium (CHROMagar, https://www.chromagar.com) and confirmed by sequencing of the ribosomal internal transcribed spacer (ITS) region. Most cases were identified in 2019; C. vulturna infections were identified in 2 patients during January 1, 2020–January 1, 2022. Enhanced hygiene measures taken at that time may have dampened the spread of C. vulturna in the hospital.
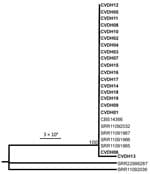
On the basis of results of the ITS and multilocus sequence typing for 8 conserved genes, we then performed phylogenetic analyses on the isolates. All strains isolated in this study (CVDH01–19) were closely related by phylogenetic analyses and clustered together in 1 clade (Figure 1; Appendix Figure 2).
The hospital has 1 intensive care unit (ICU). Of the 19 patients we identified as infected with C. vulturna, 11 were from the ICU, 4 were from the neuroscience ward, and 4 were from other departments within the hospital. The age range of patients was 13–83 years (median 63 years). Because all patients had PICC lines for delivery of medications and C. vulturna strains were isolated from the PICC line tips of 7 patients, the use of PICC lines could be a major risk factor for C. vulturna infection. Other risk factors could include traumatic injuries, hypertension, cancer, and blood and pulmonary infections (Appendix Table). We also conducted environmental screening assays but were unable to detect or isolate C. vulturna from hospital surfaces, including walls, floors, bedside tables, bed sheets, bed frames, blood pressure cuffs, and chairs.
We used 1 representative C. vulturna strain from each patient for subsequent antifungal drug susceptibility testing and phenotypic analyses (Appendix Table). Using the breakpoints established for C. albicans, we determined that all 19 of the C. vulturna strains tested were resistant to azole drugs (Table). All isolates were resistant to amphotericin B (MIC 4 mg/L) but were susceptible to echinocandins (MICs <0.125 for caspofungin, <0.125 for anidulafungin, <0.5 for micafungin), and flucytosine (MIC 0.06).
When grown in liquid media, we observed that the cells from the C. vulturna (CVDH) strains isolated in this study formed large aggregates and exhibited enhanced adhesion and biofilm formation abilities. This feature was similar to that of C. auris strain SJ01, which formed enhanced biofilms under both in vitro and in vivo conditions (14). (Figure 2; Appendix Figure 3).
A serious threat to human health is the emergence of new multidrug-resistant fungal species. Both the widespread use of antifungal agents and the reduced susceptibility of these emerging species to antifungal drugs could contribute to the epidemiologic shifts toward multidrug-resistant fungal pathogens that we are increasingly observing in clinical settings. In this study, we report an outbreak of C. vulturna, which is phylogenetically closely related to C. haemulonii and C. auris, in a general hospital in Shanxi Province, China. We observed that the implementation of general enhanced hygiene measures remarkably decreased overall infection rates during the COVID-19 pandemic period (January 1, 2020–January 1, 2022) in this hospital; our findings suggest that the transmission of C. vulturna may be preventable through enhanced disinfection methods. Most of the C. vulturna isolates we obtained were from patients with bloodstream infections, defined as a single isolation of C. vulturna from blood obtained through venipuncture. Phylogenetic analyses indicated that the outbreak strains were closely related (Figure 1; Appendix Figure 2), implying that those strains could have originated from the same ancestor.
Striking characteristics of the C. vulturna strains isolated in this study were their enhanced adhesion and biofilm formation abilities. It is conceivable that those characteristics may be key contributors in promoting the spread of C. vulturna strains between patients during this outbreak. Consistent with this hypothesis, we observed that the use of PICC lines was a critical risk factor for C. vulturna infections. Another notable characteristic of the C. vulturna strains isolated in this study was their reduced susceptibilities to azole drugs and amphotericin B (Table), which has also been observed in other species of the C. haemulonii complex (6,7,13).
The occurrence of infections caused by fungal species of the Metschnikowia clade has become more and more frequent in clinical settings, especially during 2009–2022 (1,6,8,13). The widespread use of antifungal drugs in clinical settings and fungicides in agricultural settings could be contributors to the increased emergence of these multidrug resistant fungal pathogens. Given the transmissible, adhesive, and antifungal drug–resistant characteristics of emerging C. vulturna clinical isolates, C. vulturna could be a serious upcoming threat to hospital infections worldwide.
Dr. Du is an associate professor in the State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, China. Her primary research interest is in the biology of pathogenic fungi.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (grant no. 2021YFC2300400 to G.H. and no. 2022YFC2303000 to H.D. and J.B.), National Natural Science Foundation of China (award nos. 82172290 and 82002123 to H.D., nos. 31930005 and 82272359 to G.H., nos. 32170193 and 32000018 to J.B., and no. 81960367 to P.Z.), Shanghai Municipal Science and Technology Major Project (award no. HS2021SHZX001 to G.H.), Jiangxi Provincial Natural Science Foundation (award no. 20212BAB206060 to P.Z.), the US National Institutes of Health National Institute of General Medical Sciences (grant no. R35GM124594 to C.J.N.), and by the Kamangar family in the form of an endowed chair (to C.J.N.). The content is the sole responsibility of the authors and does not represent the views of the funders. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
C.J.N. is a cofounder of BioSynesis, Inc., a company developing diagnostics and therapeutics for biofilm infections.
References
- Gade L, Muñoz JF, Sheth M, Wagner D, Berkow EL, Forsberg K, et al. Understanding the emergence of multidrug-resistant Candida: using whole-genome sequencing to describe the population structure of Candida haemulonii species complex. Front Genet. 2020;11:554. DOIPubMedGoogle Scholar
- Sipiczki M, Tap RM. Candida vulturna pro tempore sp. nov., a dimorphic yeast species related to the Candida haemulonis species complex isolated from flowers and clinical sample. Int J Syst Evol Microbiol. 2016;66:4009–15. DOIPubMedGoogle Scholar
- Zurita J, Paz Y Miño A, Solís MB, Sevillano G. Failed identification of Candida vulturna using the updated Vitek 2 yeast identification system, version 9.02 and CHROMagar Candida Plus. New Microbes New Infect. 2022;48:
101012 . DOIPubMedGoogle Scholar - Navarro-Muñoz JC, de Jong AW, Gerrits van den Ende B, Haas PJ, Then ER, Mohd Tap R, et al. The high-quality complete genome sequence of the opportunistic fungal pathogen Candida vulturna CBS 14366T. Mycopathologia. 2019;184:731–4. DOIPubMedGoogle Scholar
- Santos MA, Gomes AC, Santos MC, Carreto LC, Moura GR. The genetic code of the fungal CTG clade. C R Biol. 2011;334:607–11. DOIPubMedGoogle Scholar
- Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, et al. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis. 2009;48:e57–61. DOIPubMedGoogle Scholar
- Ramos LS, Figueiredo-Carvalho MH, Barbedo LS, Ziccardi M, Chaves AL, Zancopé-Oliveira RM, et al. Candida haemulonii complex: species identification and antifungal susceptibility profiles of clinical isolates from Brazil. J Antimicrob Chemother. 2015;70:111–5. DOIPubMedGoogle Scholar
- Hou X, Xiao M, Chen SC, Wang H, Cheng JW, Chen XX, et al. Identification and antifungal susceptibility profiles of Candida haemulonii species complex clinical isolates from a multicenter study in China. J Clin Microbiol. 2016;54:2676–80. DOIPubMedGoogle Scholar
- Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, et al.; Candida auris Incident Management Team. Candida auris: a review of the literature. Clin Microbiol Rev. 2017;31:
e00029-17 . DOIPubMedGoogle Scholar - Du H, Bing J, Hu T, Ennis CL, Nobile CJ, Huang G. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16:
e1008921 . DOIPubMedGoogle Scholar - Jackson BR, Chow N, Forsberg K, Litvintseva AP, Lockhart SR, Welsh R, et al. On the origins of a species: what might explain the rise of Candida auris? J Fungi (Basel). 2019;5:58. DOIPubMedGoogle Scholar
- Casadevall A, Kontoyiannis DP, Robert V. On the emergence of Candida auris: climate change, azoles, swamps, and birds. MBio. 2019;10:
e01397-19 . DOIPubMedGoogle Scholar - Chen XF, Zhang H, Jia XM, Cao J, Li L, Hu XL, et al. Antifungal susceptibility profiles and drug resistance mechanisms of clinical Candida duobushaemulonii isolates from China. Front Microbiol. 2022;13:
1001845 . DOIPubMedGoogle Scholar - Bing J, Guan Z, Zheng T, Zhang Z, Fan S, Ennis CL, et al. Clinical isolates of Candida auris with enhanced adherence and biofilm formation due to genomic amplification of ALS4. PLoS Pathog. 2023;19:
e1011239 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: June 12, 2023
1These first authors contributed equally to this article.
2These senior authors contributed equally to this article.
Table of Contents – Volume 29, Number 7—July 2023
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Ping Zhan, Dermatology Department, Affiliated Hospital of Jiangxi University of Chinese Medicine, Nanchang, 330006, China
Top