Technical Report: Highly Pathogenic Avian Influenza A(H5N1) Viruses
Updated March 17, 2023
The latest H5N1 technical report and all previous technical reports are available from the H5N1 technical report webpage.
An addendum to this report was posted on April 17, 2023. The addendum summarizes the case and genomic analysis of the virus from the first H5N1 infection reported in a human in Chile. The overall risk to human health associated with the ongoing A(H5N1) outbreaks in wild birds and poultry has not changed and remains low at this time.
Executive summary
Since 2022, despite the wide geographic spread of highly pathogenic avian influenza (HPAI) A(H5N1) viruses in wild birds and to poultry worldwide, with sporadic spillover to mammals, only a small number of sporadic human cases of A(H5N1) have been identified. All reported human cases since 2022 were associated with recent poultry exposures, and no cases of human-to-human transmission have been identified. To date, HPAI A(H5N1) viruses currently circulating in birds and poultry, with spillover to mammals, and those that have caused human infections do not have the ability to easily bind to receptors that predominate in the human upper respiratory tract. Therefore, the current risk to the public from HPAI A(H5N1) viruses remains low. However, because of the potential for influenza viruses to rapidly evolve and the wide global prevalence of HPAI A(H5N1) viruses in wild birds and poultry outbreaks, continued sporadic human infections are anticipated. Continued comprehensive surveillance of these viruses in wild birds, poultry, mammals, and people worldwide, and frequent reassessments are critical to determine the public health risk, along with ongoing preparedness efforts.
- CDC is actively working on the domestic situation with clade 2.3.4.4b HPAI A(H5N1) viruses in wild birds and poultry outbreaks, including conducting surveillance among people with relevant exposures and preparing for the possibility that contemporary HPAI A(H5N1) viruses gain the ability for increased transmissibility to people.
- CDC, along with our state and local public health partners, continues to actively monitor people in the United States who have been exposed to infected birds and poultry for 10 days after exposure. To date, more than 6,300 people in 52 jurisdictions have been monitored since 2022, and only one human case has been identified
- An H5 candidate vaccine virus (CVV) produced by CDC is nearly identical or, in many samples, identical to the hemagglutinin (HA) protein of recently detected clade 2.3.4.4b HPAI A(H5N1) viruses in birds and mammals (including the recent outbreak in farmed mink in Spain) and could be used to produce a vaccine for people, if needed, and would provide good protection against the clade 2.3.4.4b HPAI A(H5N1) viruses circulating in birds. This H5 CVV is available and has been shared with vaccine manufacturers.
- Because influenza viruses are constantly changing, CDC performs ongoing analyses of A(H5N1) viruses to identify genetic changes that might allow for spread more easily to and between people, cause serious illness in people, reduce susceptibility to antivirals, affect the sensitivity of diagnostic assays, or reduce neutralization of the virus by vaccine induced antibodies. To date, no such concerning changes have been identified in HPAI A(H5N1) viruses circulating in wild birds and poultry worldwide or that have sporadically infected humans.
- Currently, HPAI A(H5N1) viruses are believed to pose a low risk to the health of the general public in the United States; however, people who have job-related or recreational exposures to infected birds may be at higher risk of infection and should take appropriate precautions outlined in CDC guidance.
- Comprehensive surveillance and readiness efforts are ongoing, and CDC continually takes preparedness measures to be ready in case the risk to people from HPAI A(H5N1) or other novel influenza A viruses changes.
HPAI A(H5N1) viruses in wild birds and poultry
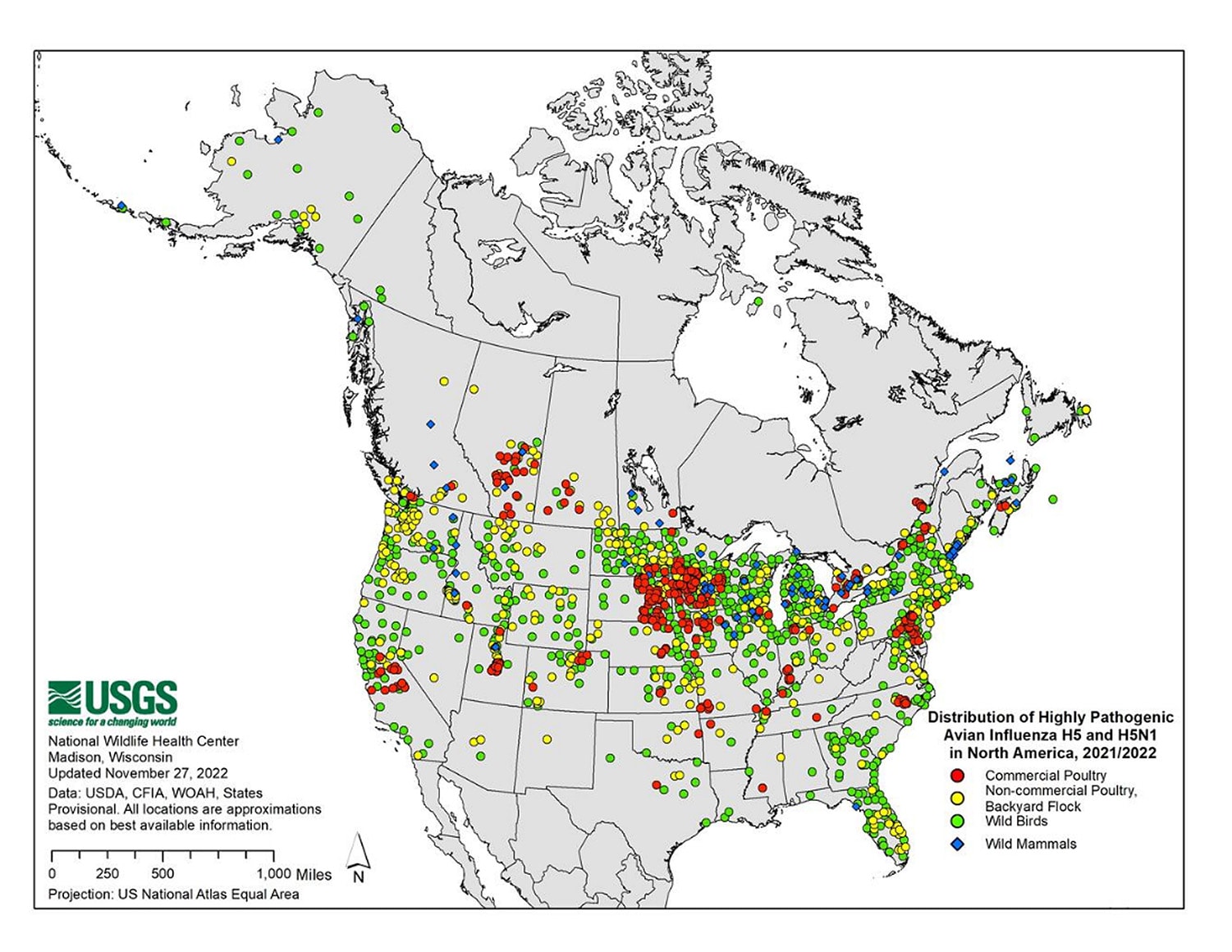
Since 2005, HPAI A(H5N1) viruses have undergone extensive genetic diversification including the formation of hundreds of genotypes following reassortment with other avian influenza A viruses. Clade 2.3.4.4b HPAI A(H5N1) viruses emerged in 2020 and were introduced into North America in late 2021 [1] and have spread to Central and South America, resulting in wild bird and poultry outbreaks in many countries [2].
Globally, this 2.3.4.4b clade of HPAI A(H5N1) viruses has become widespread causing record numbers of bird outbreaks in wild, backyard, village, and farm birds. Over 11,300 animal outbreaks of HPAI A(H5N1) viruses were reported by 73 member states to the World Organization for Animal Health since January 2022.
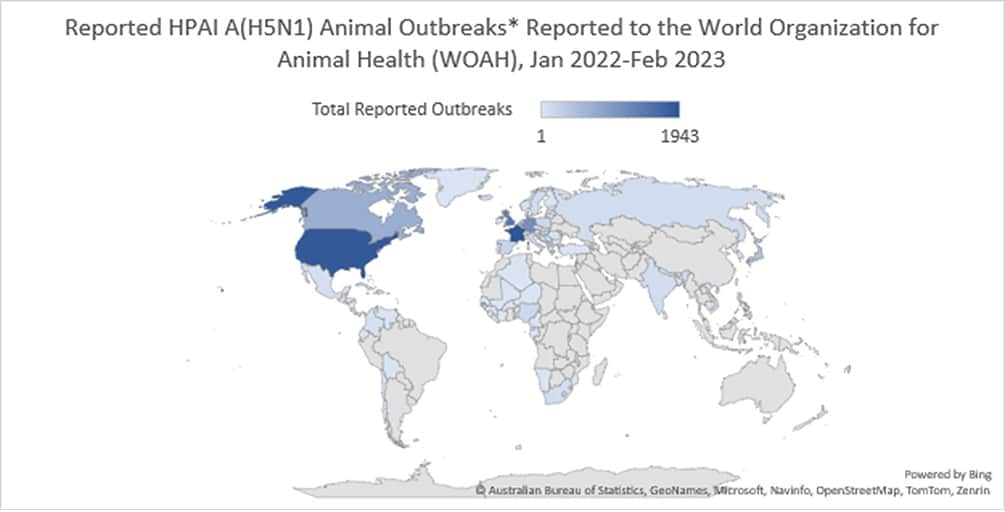
* WOAH defines an outbreak as an occurrence of one or more cases in a group of animals with a defined epidemiologic relationship therefore outbreak numbers don’t reflect the quantity of animal affected.
In the United States, USDA APHIS monitors for avian influenza viruses in wild, commercial, and backyard birds. From January 2022 to March 15, 2023, APHIS reported HPAI A(H5)/A(H5N1) virus detections in 6,444 wild birds in 49 states and 799 commercial and backyard flocks in 47 states.
HPAI A(H5N1) virus infections among mammals
Sporadic HPAI A(H5N1) virus infections of mammals have been reported for 20 years in countries that have experienced HPAI A(H5N1) virus outbreaks in poultry or wild birds. HPAI A(H5) viruses are known to occasionally infect mammals that eat (presumably infected) birds or poultry and mammals that are exposed to environments with a high concentration of virus.
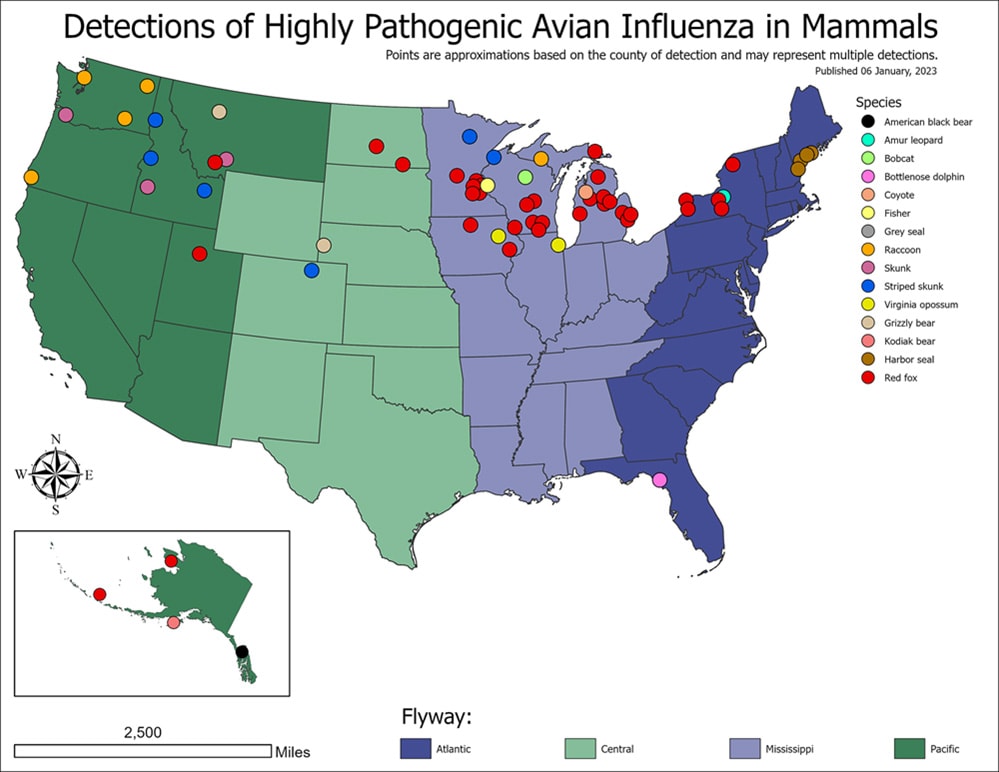
In the United States, from May 2022 to March 15, 2023, USDA APHIS reported HPAI A(H5N1) virus detections in 144 mammals in 23 states. Globally, sporadic HPAI A(H5N1) virus infections have been reported in farmed mink in Spain, sea lions in Peru and Chile, and foxes in Canada, France, and other countries. The reports of HPAI A(H5N1) virus infections in mammals are not surprising given the widespread outbreaks of HPAI A(H5N1) virus infections in wild birds.
Genetic data have revealed that when some mammals are infected with HPAI A(H5N1) virus, the virus may undergo intra-host evolution resulting in genetic changes that allow the virus to replicate more efficiently in the lower respiratory tract [3-5].
Although these genetic changes may impact mammalian disease outcome, they have not been associated with enhanced transmissibility of the virus to humans. HPAI A(H5N1) viruses do not currently have an ability to easily infect and bind to α2,6-linked sialic acid receptors that are predominant in the human upper respiratory tract [6], which would be needed to increase the risk of transmission to people [7,8].
Human cases of A(H5N1)
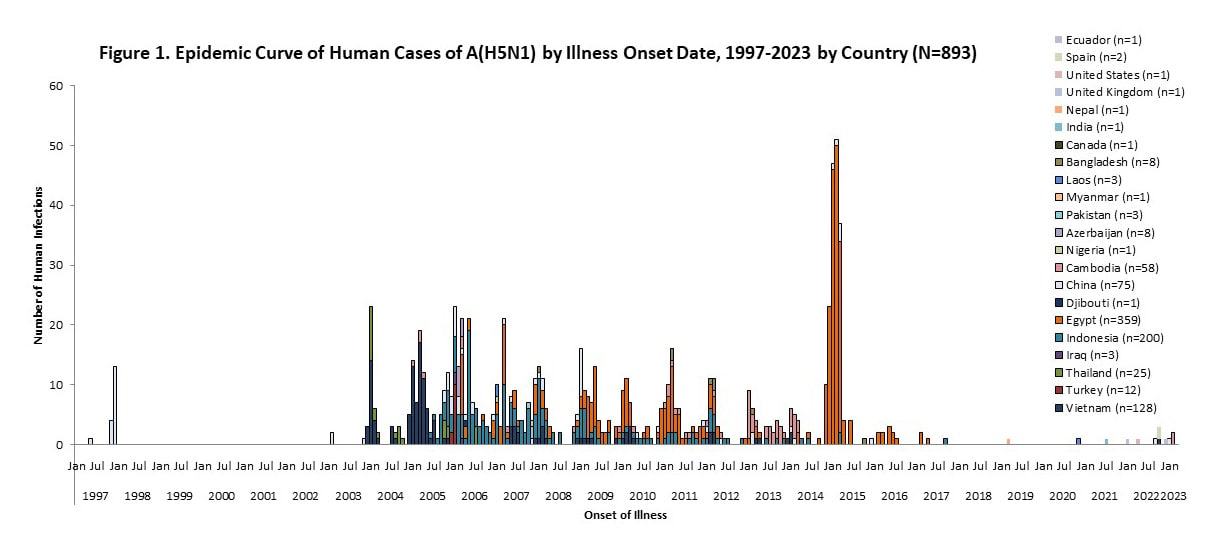
While HPAI A(H5N1) viruses are currently circulating widely in wild birds and poultry in many geographic regions, relatively few human cases of A(H5N1) have been reported in recent years [Figure 1]. Between January 2022 and March 15, 2023, ten sporadic human cases of A(H5N1) were reported from seven countries [Table 1].
One human case of A(H5N1) was reported in the United States in April 2022. The individual reported fatigue without other symptoms and a low level of A(H5N1) viral RNA was detected in a single upper respiratory tract specimen. It is possible that detection of A(H5N1) viral RNA resulted from deposition of non-infectious viral material in the upper respiratory tract of the individual and did not represent true infection, similar to the environmental contamination that was attributed to the two asymptomatic cases reported in Spain [9].
All reported cases had recent exposure to sick or dead poultry, and no cases of human-to-human HPAI A(H5N1) virus transmission were identified. Five cases (3 children, 2 adults) had severe disease, and 2 died. Seven cases were associated with clade 2.3.4.4b HPAI A(H5N1) viruses, and two cases were associated with clade 2.3.2.1c HPAI A(H5N1) viruses; none of the HPAI A(H5N1) virus genetic sequences contained any known markers of reduced susceptibility to currently recommended FDA-approved influenza antiviral medications.
| Country of Case | Month of illness onset or case detection | Disease Severity and Outcome | Virus Clade by sequencing or associated poultry outbreaks |
|---|---|---|---|
| Cambodia | February 2023 | Critical illness, died | Clade 2.3.2.1c |
| Cambodia | February 2023 | Mild illness, survived | Clade 2.3.2.1c |
| China | September 2022 | Critical illness, died | Clade 2.3.4.4b |
| China | January 2023 | Hospitalized, outcome not reported | Clade 2.3.4.4b |
| Ecuador | December 2022 | Critical illness, survived | Clade 2.3.4.4b |
| Spain | September 2022 | Asymptomatic | Clade 2.3.4.4b |
| Spain | October 2022 | Asymptomatic | Clade 2.3.4.4b |
| United Kingdom | January 2022 | Asymptomatic | Clade 2.3.4.4b |
| United States | April 2022 | Fatigue only, survived | Clade 2.3.4.4b |
| Vietnam | October 2022 | Critical illness, survived | Not reported |
Since 1997, a total of 893 sporadic human A(H5N1) cases have been reported from 21 countries [Figure 1], caused by different HPAI A(H5N1) virus clades [10], with a cumulative case fatality proportion of greater than 50%. Human A(H5N1) cases peaked in 2006 (115 cases, 9 countries) and 2015 (145 cases, 4 countries) primarily due to a large epidemic in Egypt with 136 cases [Figure 1].
Nearly all reported human A(H5N1) cases had poultry exposures, such as to sick or dead poultry or visiting live poultry markets. Rare probable, limited, non-sustained human-to-human HPAI A(H5N1) influenza virus transmission likely occurred in a small number of family members following prolonged, close unprotected exposure with a symptomatic case-patient during 2004-2007 in multiple countries [11-14].
Active monitoring of persons exposed to HPAI A(H5N1) virus in the United States
Although few human cases have occurred recently, given widespread infection among poultry and wild birds, people who have job-related or recreational exposures to infected birds or sick or dead mammals might be at higher risk of infection.
CDC, in collaboration with state, territorial, and local public health partners, actively monitors people exposed to infected birds and poultry for 10 days after their last exposure. As of March 15, 2023:
- Total monitored: more than 6,300 people in 52 jurisdictions since February 2022
- Total illnesses reported among monitored persons: more than 160 people
- Number positive for influenza A(H5N1) virus: 1
Of the approximately 160 people showing symptoms who were tested for novel influenza A and seasonal flu viruses along with other respiratory viruses, HPAI A(H5N1) virus genetic material was detected in a respiratory specimen from one person in Colorado who experienced fatigue without any other symptoms while participating in poultry culling activities. [See above section on “Human cases of A(H5N1).”]
U.S. influenza surveillance for human infections with novel influenza A viruses, including HPAI A(H5N1) virus
Human infection with a novel influenza A virus, including HPAI A(H5N1) virus, is a nationally notifiable condition (case definition: Novel Influenza A Virus Infections 2014 Case Definition | CDC)
Influenza testing is widely available in clinical laboratories and healthcare facilities. Assays in these settings would detect A(H5N1) virus infections as influenza A positive and a subset of assays would be able to also determine that they are not currently circulating seasonal influenza A virus subtypes (H1 or H3). Specimens from persons possibly exposed to H5N1 virus or that test positive for an influenza A virus but negative for A(H1) and A(H3) subtypes should be forwarded to the appropriate state or local public health laboratory for further testing. Very few specimens have been submitted to CDC for H5 testing since January 2022.
- Seasonal influenza virus detection assays that can also detect novel influenza A viruses are used in 128 public health laboratories in all 50 U.S states.
- Specific diagnostic assays to detect current A(H5) viruses are available at 99 public health laboratories in all 50 states.
Per long-standing standard protocols, upon detection of a virus that tests positive for H5, or a virus that cannot be subtyped as currently circulating H1 or H3, the public health laboratory will contact CDC and ship the specimen to CDC for confirmation. An investigation of the case will be initiated, and a case report form submitted to CDC through the novel influenza A reporting module.
Activity
Activity
Summary
Summary
Global surveillance and rapid response to human infections
Global surveillance and rapid response to human infections
| CDC’s Influenza Division supports surveillance in live bird markets, backyard farms, and wild birds and/or their environments in Bangladesh, Cambodia, China, Guatemala, Kenya, Lao PDR, Peru, Thailand, and Vietnam. Surveillance data highlight the high prevalence and wide range of avian influenza A viruses in birds and help to describe the changing epidemiology of avian influenza A viruses.
In 2022, the Influenza Division tracked more than 50 human infections with avian influenza A viruses reported to the WHO from seven countries in four WHO regions. Most recently, CDC Influenza Division field staff assisted in the rapid response investigations of two human H5N1 cases in Cambodia in 2023. |
| CDC’s Influenza Division supports surveillance in live bird markets, backyard farms, and wild birds and/or their environments in Bangladesh, Cambodia, China, Guatemala, Kenya, Lao PDR, Peru, Thailand, and Vietnam. Surveillance data highlight the high prevalence and wide range of avian influenza A viruses in birds and help to describe the changing epidemiology of avian influenza A viruses.
In 2022, the Influenza Division tracked more than 50 human infections with avian influenza A viruses reported to the WHO from seven countries in four WHO regions. Most recently, CDC Influenza Division field staff assisted in the rapid response investigations of two human H5N1 cases in Cambodia in 2023. |
Virological assessments
Virological assessments
Because influenza viruses have a high error rate during replication and rapidly evolve, CDC continually conducts genetic analyses of viruses to identify changes that may impact virus phenotypes such as antigenicity, antiviral susceptibility, transmissibility, and/or pathogenesis. Genetic analysis is also performed to assess changes that may impact diagnostic test performance.
Because influenza viruses have a high error rate during replication and rapidly evolve, CDC continually conducts genetic analyses of viruses to identify changes that may impact virus phenotypes such as antigenicity, antiviral susceptibility, transmissibility, and/or pathogenesis. Genetic analysis is also performed to assess changes that may impact diagnostic test performance.
Diagnostics
Diagnostics
Various CDC influenza virus diagnostic real time RT-PCR tests detect typical human (seasonal) viruses or novel influenza A viruses (e.g., H5, H7) that may infect people through zoonotic transmission. These diagnostic tests are used in all 50 U.S states and globally. Additionally, there are CDC diagnostic tests that specifically detect the current H5 viruses, which are available in public health laboratories in all 50 U.S. states and international laboratories.
Most commercial assays used for human influenza virus testing are likely to detect HPAI A(H5N1) viruses because they target conserved proteins.
Various CDC influenza virus diagnostic real time RT-PCR tests detect typical human (seasonal) viruses or novel influenza A viruses (e.g., H5, H7) that may infect people through zoonotic transmission. These diagnostic tests are used in all 50 U.S states and globally. Additionally, there are CDC diagnostic tests that specifically detect the current H5 viruses, which are available in public health laboratories in all 50 U.S. states and international laboratories.
Most commercial assays used for human influenza virus testing are likely to detect HPAI A(H5N1) viruses because they target conserved proteins.
Candidate vaccine virus development
Candidate vaccine virus development
The development of influenza candidate vaccine viruses (CVVs), coordinated by WHO, remains an essential component of the overall global strategy for influenza pandemic preparedness. A library of H5 candidate vaccine viruses (CVV) has been produced with additional recommendations for development during bi-annual vaccine consultation meetings (See Table and https://www.who.int/teams/global-influenza-programme/vaccines/who-recommendations/zoonotic-influenza-viruses-and-candidate-vaccine-viruses). The CDC Influenza Risk Assessment Tool is also used to help prioritize HPAI A(H5) viruses for development of CVVs.
A/Astrakhan/3212/2020-like and A/American wigeon/South Carolina/22-000345-001/2021-like CVVs closely related HPAI A(H5N1) (clade 2.3.4.4b) viruses circulating in North America have been developed and are available for vaccine manufacturers. The two CVVs produced by the U.S. CDC (i.e., IDCDC-RG71A and IDCDC-RG78A) and one CVV produced by U.S. FDA (CBER-RG8A) encode hemagglutinin (HA) proteins that are nearly identical or identical to the HA of most recent clade 2.3.4.4b H5N1 viruses detected in birds and mammals and could be used to produce a vaccine for people if needed. One additional clade 2.3.4.4b H5N1 CVV has been recommended for development as part of pandemic preparedness.
The development of influenza candidate vaccine viruses (CVVs), coordinated by WHO, remains an essential component of the overall global strategy for influenza pandemic preparedness. A library of H5 candidate vaccine viruses (CVV) has been produced with additional recommendations for development during bi-annual vaccine consultation meetings (See Table and https://www.who.int/teams/global-influenza-programme/vaccines/who-recommendations/zoonotic-influenza-viruses-and-candidate-vaccine-viruses). The CDC Influenza Risk Assessment Tool is also used to help prioritize HPAI A(H5) viruses for development of CVVs.
A/Astrakhan/3212/2020-like and A/American wigeon/South Carolina/22-000345-001/2021-like CVVs closely related HPAI A(H5N1) (clade 2.3.4.4b) viruses circulating in North America have been developed and are available for vaccine manufacturers. The two CVVs produced by the U.S. CDC (i.e., IDCDC-RG71A and IDCDC-RG78A) and one CVV produced by U.S. FDA (CBER-RG8A) encode hemagglutinin (HA) proteins that are nearly identical or identical to the HA of most recent clade 2.3.4.4b H5N1 viruses detected in birds and mammals and could be used to produce a vaccine for people if needed. One additional clade 2.3.4.4b H5N1 CVV has been recommended for development as part of pandemic preparedness.
Vaccines
Vaccines
Influenza virus strains of pandemic potential change over time and multiple new strains circulate in animals every year without leading to sustained human-to-human transmission. The U.S. government has a preparedness program that enables a rapid response to influenza virus strains as the strains evolve. As part of this program, the Biomedical Advanced Research and Development Authority (BARDA) works with private industry partners to make and test small quantities of updated vaccines that match new strains of influenza viruses with pandemic potential as they emerge in case any of them result in sustained human-to-human transmission, while at the same time, supporting manufacturing capacity to allow for large-scale vaccine production when needed.
Influenza virus strains of pandemic potential change over time and multiple new strains circulate in animals every year without leading to sustained human-to-human transmission. The U.S. government has a preparedness program that enables a rapid response to influenza virus strains as the strains evolve. As part of this program, the Biomedical Advanced Research and Development Authority (BARDA) works with private industry partners to make and test small quantities of updated vaccines that match new strains of influenza viruses with pandemic potential as they emerge in case any of them result in sustained human-to-human transmission, while at the same time, supporting manufacturing capacity to allow for large-scale vaccine production when needed.
Limitations of the Report
This report is subject to the following limitations. First, the number of human infections with currently circulating clade 2.3.4.4b HPAI A(H5N1) viruses is small. Conclusions regarding virus characterization analyses, transmissibility from animals to people, transmissibility among people, and clinical spectrum of illness in people should be interpreted in light of this small number. Second, detailed exposure information was not available for all persons actively monitored for illness after exposure to HPAI A(H5N1) virus-infected birds and poultry in the United States. Thus, we were not able to assess the impact of exposure variables such as duration of exposure, nature of exposure (e.g., direct vs. indirect contact), and use of personal protective equipment on infection risk.
Conclusions
- To date, CDC analyses of clade 2.3.4.4b HPAI A(H5N1) viruses detected in wild birds, poultry, and sporadically in mammals since late 2021 indicate that these viruses all have a high degree of genetic identity with each other and no significant mammalian adaptive substitutions, insertions or deletions have been identified, particularly in the HA gene, which is important for zoonotic and subsequent human-to-human transmission.
- Considering the high prevalence of HPAI A(H5N1) viruses detected in wild birds, and poultry worldwide, spill over into mammals (particularly carnivores that may feed on infected avian species), additional sporadic zoonotic infections among people with exposures to sick or dead poultry or wild birds are anticipated.
- HA clade 2.3.4.4b A(H5N1) viruses currently circulating in wild birds and poultry worldwide lack the ability to preferentially bind to the types of sialic acid receptors that are predominant in the upper respiratory tract of humans and therefore do not currently have the ability to easily infect or transmit among people.
- Despite extensive worldwide spread of influenza A(H5N1) viruses in wild birds and poultry in recent years, only a small number of sporadic human infections with 2.3.4.4b or clade 2.3.2.1c H5N1 viruses have been reported since 2022; all cases had recent exposure to poultry and no cases of human-to-human influenza A(H5N1) virus transmission were identified.
While CDC’s assessment is that the overall threat of HA clade 2.3.4.4b A(H5N1) viruses to public health is currently low, the widespread geographic prevalence of infected birds and poultry raises the potential for exposures of humans and other mammals that could result in viral evolution or reassortment events which might change the current risk assessment. Vigilance and ongoing surveillance of HPAI A(H5N1) viruses circulating in wild birds, poultry, and in sporadic infections of mammals and people worldwide is critical to monitor the public health risk and to detect genetic changes (particularly in the HA gene) that would change CDC’s risk assessment.
References
- Bevins SN, Shriner SA, Cumbee JC Jr, Dilione KE, Douglass KE, Ellis JW et al. Intercontinental Movement of Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4 Virus to the United States, 2021. Emerg Infect Dis. 2022 May;28(5):1006-1011.
- World Health Organization. Antigenic and genetic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness. February 2023. https://cdn.who.int/media/docs/default-source/influenza/who-influenza-recommendations/vcm-northern-hemisphere-recommendation-2023-2024/20230224_zoonotic_recommendations.pdf?sfvrsn=38c739fa_4 [1.17 MB, 12 pages]
- Gabriel G, Czudai-Matwich V, Klenk HD. Adaptive mutations in the H5N1 polymerase complex. Virus Res. 2013 Dec 5;178(1):53-62. doi: 10.1016/j.virusres.2013.05.010.
- Bogs J, Kalthoff D, Veits J, Pavlova S, Schwemmle M, Mänz B et al. Reversion of PB2-627E to -627K during replication of an H5N1 Clade 2.2 virus in mammalian hosts depends on the origin of the nucleoprotein. J Virol. 2011 Oct;85(20):10691-8. doi: 10.1128/JVI.00786-11.
- Agüero M, Monne I, Sánchez A, Zecchin B, Fusaro A, Ruano MJ et al. Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022. Euro Surveill. 2023 Jan;28(3):2300001. doi: 10.2807/1560-7917.ES.2023.28.3.2300001.
- Kandeil A, Patton C, Jones J, Jeevan T, Harrington W, Trifkovic S et al. Rapid evolution of A(H5N1) influenza viruses after intercontinental spread to North America. Research Square; 2022. DOI: 10.21203/rs.3.rs-2136604/v1.
- van Riel D, den Bakker MA, Leijten LM, Chutinimitkul S, Munster VJ, de Wit E et al. Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am J Pathol. 2010 Apr;176(4):1614-8. doi: 10.2353/ajpath.2010.090949.
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006 Mar 23;440(7083):435-6. doi: 10.1038/440435a.
- Aznar E, Casas I, González Praetorius A, Ruano Ramos MJ, Pozo F, Sierra Moros MJ et al.Influenza A(H5N1) detection in two asymptomatic poultry farm workers in Spain, September to October 2022: suspected environmental contamination. Euro Surveill. 2023 Feb;28(8):2300107. doi: 10.2807/1560-7917.ES.2023.28.8.2300107. https://pubmed.ncbi.nlm.nih.gov/36820643/
- Lai S, Qin Y, Cowling BJ, Ren X, Wardrop NA, Gilbert M et al. Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997-2015: a systematic review of individual case data. Lancet Infect Dis. 2016 Jul;16(7):e108-e118. doi: 10.1016/S1473-3099(16)00153-5.https://pubmed.ncbi.nlm.nih.gov/27211899/
- Ungchusak K, Auewarakul P, Dowell SF, Kitphati R, Auwanit W, Puthavathana P et al. Probable person-to-person transmission of avian influenza A (H5N1). N Engl J Med. 2005 Jan 27;352(4):333-40. doi: 10.1056/NEJMoa044021.https://pubmed.ncbi.nlm.nih.gov/15668219/
- Wang H, Feng Z, Shu Y, Yu H, Zhou L, Zu R et al. Probable limited person-to-person transmission of highly pathogenic avian influenza A (H5N1) virus in China.Lancet. 2008 Apr 26;371(9622):1427-34. doi: 10.1016/S0140-6736(08)60493-6.https://pubmed.ncbi.nlm.nih.gov/18400288/
- WHO Disease Outbreak News. 2006 – Indonesia. 31 May 2006. Accessed at: https://www.who.int/emergencies/disease-outbreak-news/item/2006_05_31-en
- World Health Organization. Human cases of avian influenza A (H5N1) in North-West Frontier Province, Pakistan, October-November 2007. Wkly Epidemiol Rec. 2008 Oct 3;83(40):359-64. Accessed at: https://reliefweb.int/report/guinea/weekly-epidemiological-record-wer-3-october-2008-vol-83-no-40-pp-357-364-enfr
- Technical Report: Highly Pathogenic Avian Influenza A(H5N1) Viruses (cdc.gov) – October 27, 2023
- Technical Report: Highly Pathogenic Avian Influenza A(H5N1) Viruses (cdc.gov) – October 5, 2023
- Technical Report: Highly Pathogenic Avian Influenza A(H5N1) Viruses (cdc.gov) – July 7, 2023
- Addendum: Human Infection with highly pathogenic avian influenza A(H5N1) virus in Chile (cdc.gov) – April 17, 2023
- Technical Report: Highly Pathogenic Avian Influenza A(H5N1) Viruses (cdc.gov) – March 17, 2023
- Bird Flu Current Situation Summary | Avian Influenza (Flu) (cdc.gov)
- Novel Influenza A Virus Infections (cdc.gov): An interactive dashboard of all novel influenza A virus infections in humans reported in the United States since 2010
- Reported Human Infections with Avian Influenza A Viruses
- Past Examples of Probable Limited, Non-Sustained, Person-to-Person Spread of Avian Influenza A Viruses
- Highlights in the History of Avian Influenza (Bird Flu) Timeline – 2020-2023
- Information for People Exposed to Birds Infected with Avian Influenza Viruses
- Prevention and Antiviral Treatment of Bird Flu Viruses in People
- Recommendations for Worker Protection and Use of Personal Protective Equipment (PPE) to Reduce Exposure to Novel Influenza A Viruses Associated with Severe Disease in Humans
- CDC Health Advisory, April 29, 2022 – Highly Pathogenic Avian Influenza A(H5N1) Virus: Recommendations for Human Health Investigations and Response
- Public Health Monitoring Plan for USDA/APHIS Responders to Detections of Avian Influenza Virus in Poultry [353 KB, 18 pages]
References to non-CDC sites are provided as a service and do not constitute or imply endorsement of these organizations or their programs by CDC or the U.S. Department of Health and Human Services. CDC is not responsible for the content of pages found at these sites. URL addresses listed were current as of the date of publication.