Key points
- Bat flu refers to influenza A viruses found in bats.
- In their current form, bat influenza viruses do not appear to pose a threat to human health.
- The bat species currently known to carry bat influenza viruses are not native to the continental United States, but they are common in Central and South America.

Causes and public health implications
Experts from CDC and Universidad del Valle (University of the Valley) discovered influenza A in bats during a 2009-2010 study in Guatemala. They first found influenza A in bats in little yellow-shouldered bats.1
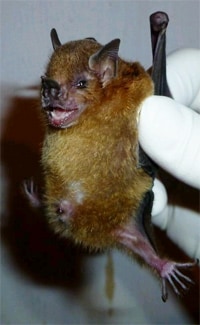
Bat influenza viruses have since been detected in some other species of bats in Central and South America.2
The discovery of bat influenza viruses is important for public health because bats represent a new animal species that may act as a source of influenza viruses. Influenza viruses are already known to spread and cause disease in other animals.
Laboratory research conducted at CDC and elsewhere suggests that bat influenza viruses would need to change significantly to be able to infect and spread easily among humans.
Risk of bat flu spreading to humans
CDC laboratory research suggests bat influenza viruses would need to undergo significant changes to become capable of infecting and spreading easily among people. Therefore, these viruses do not appear to pose a risk to human health in their current form. However, CDC scientists cannot rule out the possibility that these viruses could eventually become capable of infecting humans. The internal genes of bat influenza viruses are compatible with human influenza viruses. That means it is possible that these viruses could exchange genetic information with human influenza viruses through a process called "reassortment."
Reassortment occurs when two or more influenza viruses infect a single host cell, which allows the viruses to swap genetic information.
Reassortment facts
The conditions needed for reassortment between human influenza viruses and bat influenza viruses remain unknown. A different animal (such as pigs, horses, dogs, or seals) would need to serve as a "bridge." That means that another animal would need to be capable of being infected with both bat influenza virus and human influenza viruses for reassortment to occur.
Since the discovery of bat flu, there has been at least one study to assess the possibility of reassortment events occurring between bat influenza and other influenza viruses.3
So far, the results of these studies continue to indicate that bat influenza viruses are very unlikely to reassort with other influenza viruses. Preliminary laboratory research at CDC suggests that human cells do not support growth of bat influenza viruses.1
Difference between bat influenza viruses and other influenza viruses
The bat influenza viruses discovered in Central and South America are very different from other influenza viruses in humans and animals. All influenza A viruses have hemagglutinin (HA) surface proteins. Until the discovery of bat influenza viruses, there were only 16 different classes (or “subtypes”) of HA proteins known to exist in nature.
The new bat influenza viruses found in Central and South America are distinct from pre-existing subtypes. CDC scientists have classified them as new subtypes, denoted as “H17” and “H18.”12
The other surface protein-coding gene of bat influenza viruses, neuraminidase (NA), is different from that of known influenza viruses, as well. CDC scientists have proposed new designations for the NA subtypes found in bats: “N10” and “N11.”
Learn more about influenza viruses
Impact on influenza virus evolution
The discovery of bat flu has shed light on the evolution of influenza A, B, and C viruses. It is possible that many of the internal genes of bat influenza viruses are descendants of influenza virus families that once circulated more widely in previous centuries and that are now extinct, or yet to be discovered.
Using a process called phylogenic analysis, comparisons between different bat influenza viruses found in Central and South America have shown that there is considerable genetic diversity between them. Some researchers concluded that in Central and South American bat populations, the bat influenza viruses may have as much genetic diversity as those found in the influenza viruses of all other mammal and bird species combined.
These conclusions suggest that bat influenza viruses have been evolving in bats over a very long time, potentially centuries.2
How bat flu was discovered
The discovery of the bat influenza viruses was made possible by field work conducted by the following:
- CDC's Global Disease Detection (GDD) Regional Center in Guatemala, in collaboration with CDC's Pathogen Discovery Program.
- CDC's Rabies program.
- Universidad del Valle in Guatemala.
A total of 316 healthy bats from 21 different species were captured from eight locations in southern Guatemala during 2009 and 2010. Rectal swabs and tissues were collected from each of the bats.
Videos showing how specimens are collected from bats in Guatemala are available: "Global Disease Detectives" and "CDC Global Disease Detectives: Clues From a Bat Cave."
Swabs that were negative for rabies were analyzed by the Pathogen Discovery Program in CDC's Division of Viral Diseases to detect other pathogenic viruses. Initial viral screening determined that 3 of the 316 bats tested positive for influenza. All were from a single species known as little yellow-shouldered bats.
The Pathogen Discovery Program was able to identify and sequence the complete genome of the new viruses using high-throughput sequencing instruments in the laboratory.
- Suxiang Tong et al. "A distinct lineage of influenza A virus from bats." PNAS 2012.
- Suxiang Tong et al. "New World Bats Harbor Diverse Influenza A Viruses." PLOS Pathogens. 2013.
- M Juozapaitis et al. "An infectious bat-derived chimeric influenza virus harbouring the entry machinery of an influenza A virus." Nature Communications. 2014. 5:4448.
