What to know
This case study from Arizona provides insight into how SARS-CoV-2 sequencing is used to describe the genomic epidemiology of a state and as an investigative tool in COVID-19 outbreak settings.
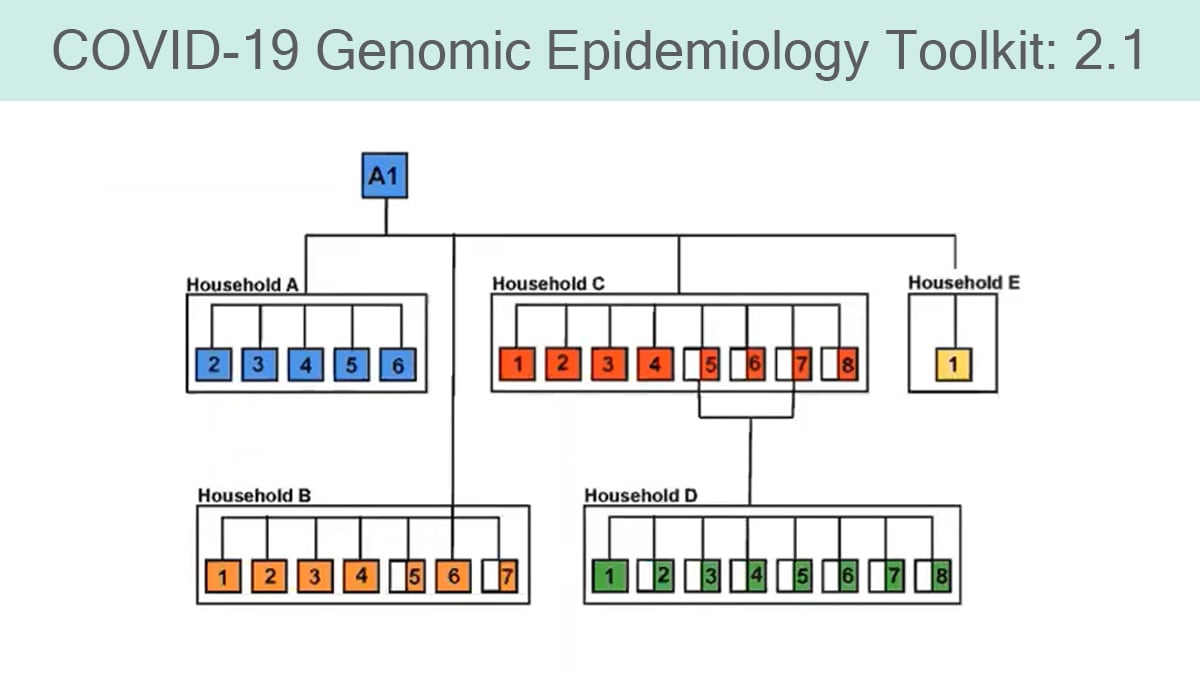
Overview
Presenter: Hayley Yaglom, MS, MPH | Genomic Epidemiologist, Translational Genomics Research Institute
View Presentation [Full Version] | [Short Version]
You can read more about this work: An Early Pandemic Analysis of SARS-CoV-2 Population Structure and Dynamics in Arizona
Further Reading
- An early pandemic analysis of SARS-CoV-2 population structure and dynamics in Arizona. Ladner, et al. mBio, 2020.
- AZ-Strain: Genomic epidemiology of SARS-CoV-2 in Arizona. Arizona COVID Genomics Union (ACGU).
Additional Resources
- Communicating results using narratives.external iconNextstrain.org (documentation).
- Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States.external iconFauver, et al. Cell, 2020.
- Large-scale sequencing of SARS-CoV-2 genomes from one region allows detailed epidemiology and enables local outbreak management.external iconPage, et al. Microb Genom, 2021.
- Viral genomes reveal patterns of the SARS-CoV-2 outbreak in Washington State.external iconMueller, et al. Sci Transl Med, 2021.
- Revealing fine-scale spatiotemporal differences in SARS-CoV-2 introduction and spread.external iconMoreno, et al. Nat Commun, 2020.
