What to know
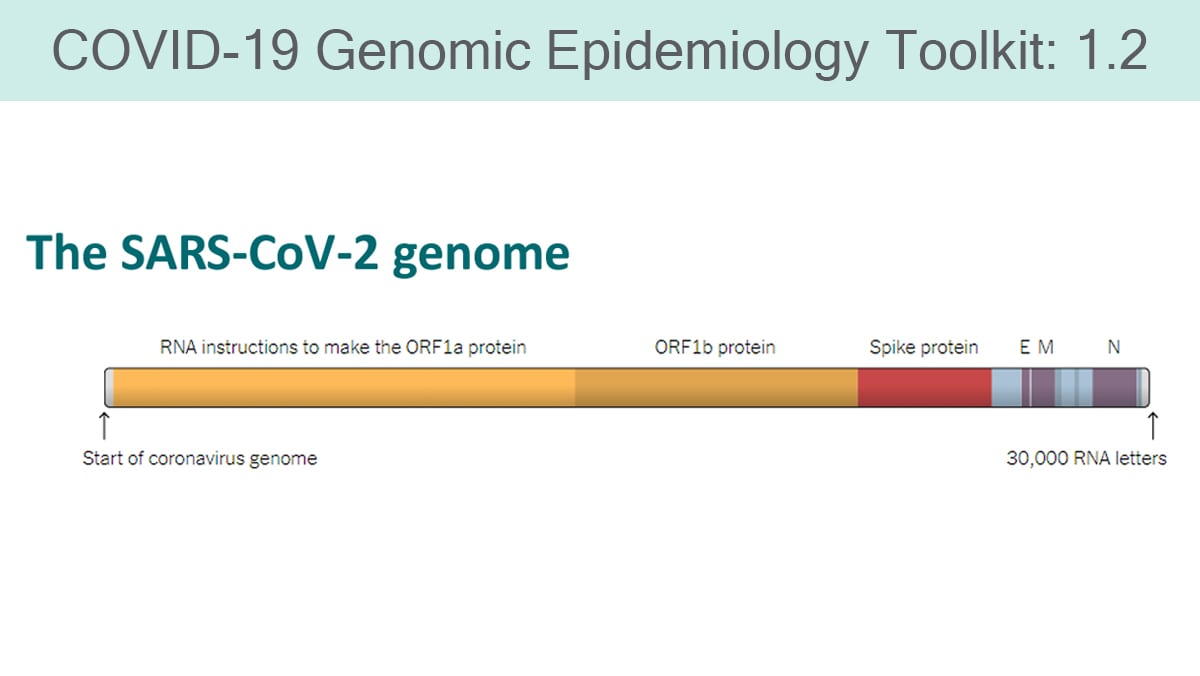
This module describes the basics of microbial genomes, with specific refence to the SARS-CoV-2 genome. Genomic sequencing and phylogenetics can be used to monitor the emergence of important new strains.
Module 1.2
Presenter: Shatavia S. Morrison, PhD | Bioinformatics Unit Lead, CDC
Download Presentation Slides [PDF – 15 slides]
Further Reading
- How coronavirus mutates and spreads.Corum and Zimmer. New York Times, 2020.
- SARS-CoV-2 sequencing data: The devil is in the genomic details.Hemarajata. ASM.org, 2020.
Additional Resources
- A pneumonia outbreak associated with a new coronavirus of probable bat origin.Zhou, et al. Nature, 2020.
- A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology.Rambaut, et al. Nat Microbiol, 2020.
- Addendum: A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology.Rambaut, et al. Nat Microbiol, 2021.
- The SARS-CoV-2 genome: variation, implication and application. [PDF, 166K]The Royal Society, 2020.
- Profiling SARS-CoV-2 mutation fingerprints that range from the viral pangenome to individual infection quasispecies.Lau, et al. Genome Med, 2021.
